Research Interests
Host-pathogen interactions
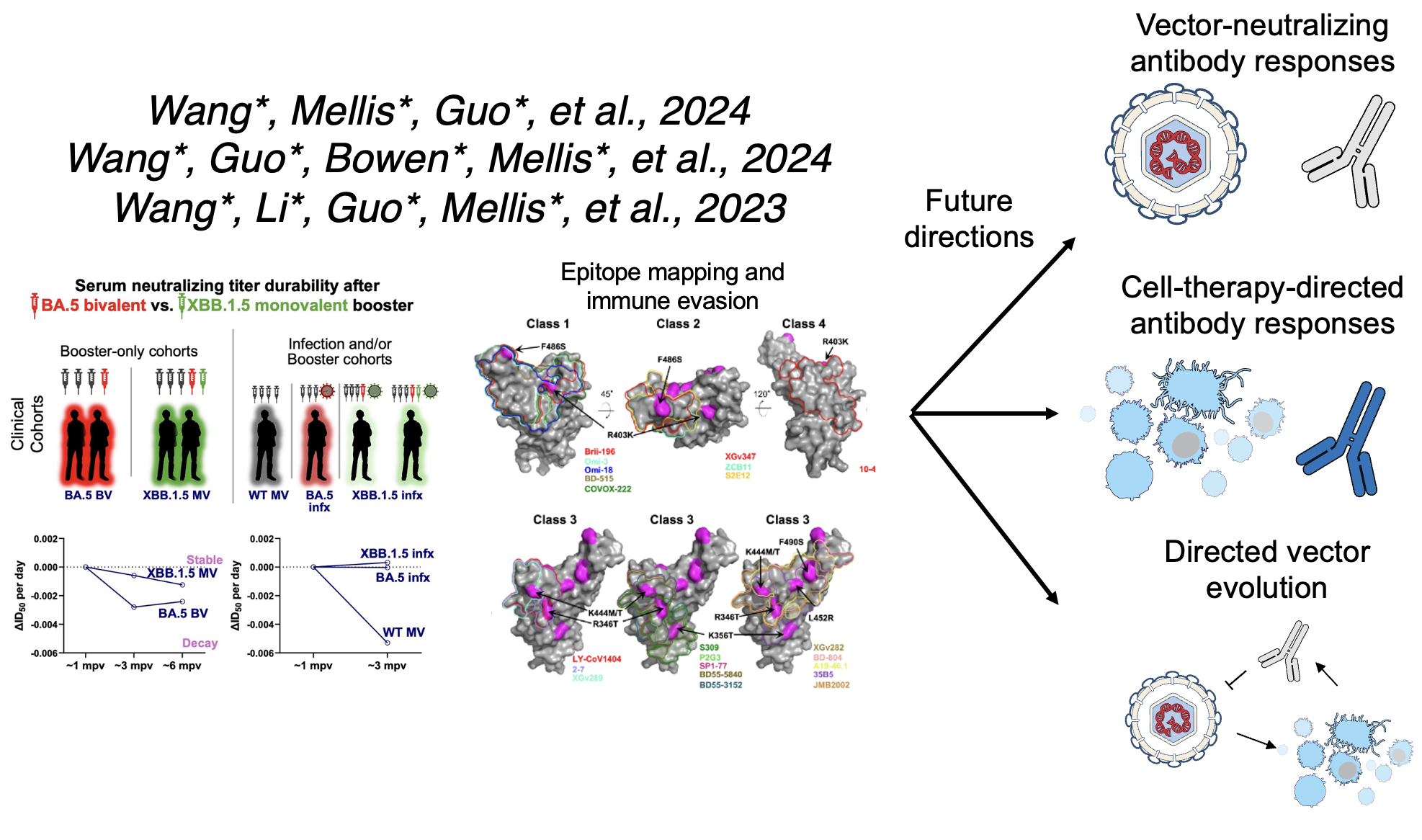
In my postdoc with Dr. David Ho, I have pursued several projects related to adaptive immune responses to viruses. In particular, I am interested in 1) better understanding neutralizing antibody responses to a variety of viruses; and 2) developing treatments (or vaccines) for viral infections using strategies informed by models of antigenic evolution and viral gene regulation. We have published or posted a number of studies about SARS-CoV-2 evolution, COVID-19 vaccines, and associated adaptive immune responses, with more on the way. Currently, I am also exploring new projects on topics including correlates of protection from SARS-CoV-2, latent HIV reservoirs, and treatments for other viruses of pandemic potential. Ultimately, I will apply insights from these studies to the field of gene and cell therapies, as well.
Gene regulation and genetic engineering
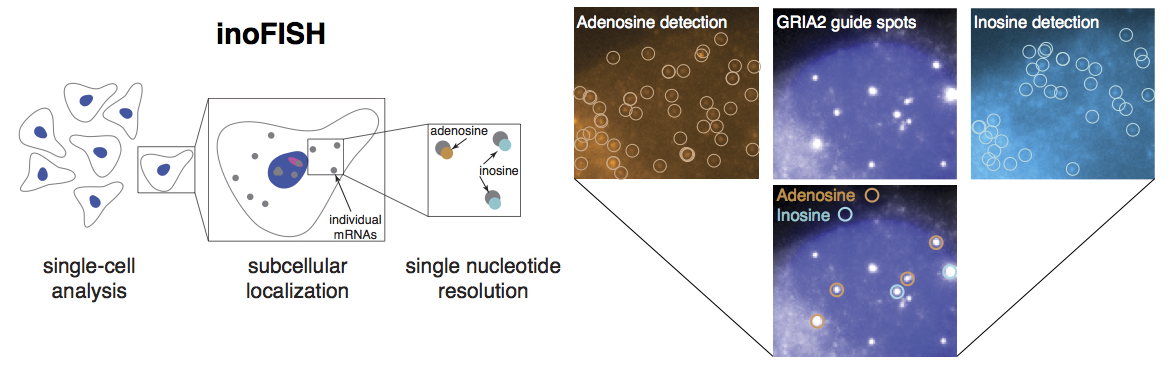
I am interested in studying gene regulation and developing related new experimental methods to answer challenging research questions. In the first part of my PhD thesis we invented a single-molecule RNA FISH-based tool, inoFISH, for visualizing RNA editing with sub-single-cell resolution. I further contributed to the development of other RNA FISH-based methods for amplifying RNA FISH signal and for visualizing allelic expression in mouse tissues. Prior to graduate school I also contributed to the design and implementation of a conditional knockout module for genome engineering. More recently, we published a study about transcriptional adaptation, a gene regulatory phenomenon that allows organisms to compensate for mutations. I am pursuing projects related to leveraging transcriptional adaptation for engineering gene upregulation in my postdoctoral research.
Systems biology of cellular identity
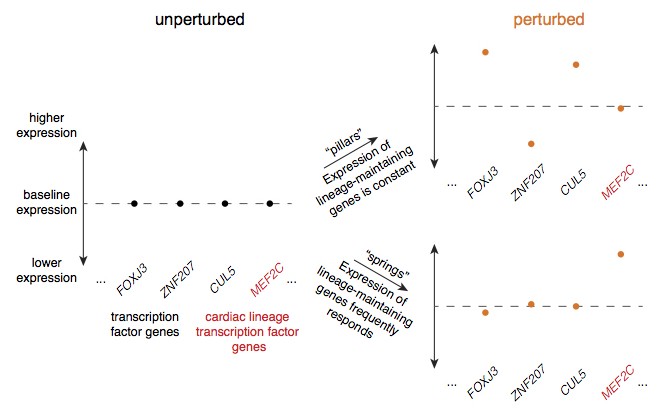
I am also interested in developing quantitative models for thinking about how and why cells have cellular identity (a.k.a. cell type) at a molecular level and from an evolutionary perspective. In my PhD thesis we identified gene regulatory trends associated with cellular identity maintenance. Relatedly, I am interested in designing protocols for reprogramming and transdifferentiation (directing cells to progenitor or other differentiated types, respectively) based on new models of the molecular and genetic basis of different cellular identities. We made progress toward this goal for more efficient fibroblast-to-iPSC reprogramming by suppressing genes that may be needed to maintain fibroblast identity. For more information, please see our paper. Currently, I am independently pursuing analytical projects about 1) how cells make tradeoffs in single-cell gene expression patterns to balance regulation of their diverse functions and 2) how to visualize cellular identity landscapes based on models of transcription factor-regulated cellular identity.